Bridging the Gap: Making OLake's MOR Iceberg Tables Compatible with Databrick's Query Engine
If you're using OLake to replicate database changes to Apache Iceberg and Databricks for analytics, you've probably hit a frustrating roadblock: Databricks doesn't support equality delete files. OLake writes data efficiently using Merge-on-Read (MOR) with equality deletes for CDC operations, but when you try to query those tables in Databricks, the deletions, updates and inserts simply aren't honored. Your query results become incorrect, missing critical data changes.
This isn't just a Databricks limitation—several major query engines including Snowflake face the same challenge. While these platforms are incredibly powerful for analytics, their Iceberg implementations only support Copy-on-Write (COW) tables or position deletes at best.
In this blog, I'll walk you through the problem and show you how we've solved it with a simple yet powerful MOR to COW write script that transforms OLake's MOR tables into COW-compatible tables that Databricks and other query engines can read correctly.
The Problem: MOR vs COW in the Real World
Let's understand what's happening under the hood. When you use OLake for Change Data Capture (CDC), it writes data to Iceberg using a strategy called Merge-on-Read (MOR) with equality delete files. This approach is optimized for high-throughput writes:
Note: For a deeper understanding of MOR vs COW strategies in Apache Iceberg, refer to our detailed guide on Merge-on-Read vs Copy-on-Write in Apache Iceberg.
How OLake Writes Data (MOR with Equality Deletes):
1. Initial Full Refresh: OLake performs a complete historical load of your table to Iceberg. This creates append only data files (No MOR).
2. CDC Updates: As changes happen in your source database, OLake captures them and creates equality delete files and data files.
3. The Result: After multiple CDC sync cycles, you have:
- Multiple data files with your records
- Multiple equality delete files tracking which records should be ignored
The Solution: Automated MOR to COW Write
The solution is to periodically compact your MOR tables into COW format—essentially creating a clean mirror table where all deletes and updates are fully applied by rewriting the data files. Think of it as "resolving" all the pending changes into a single, clean table state.
Once the COW table is created and verified, you can expire the MOR table data. To manage storage efficiently, run Iceberg's snapshot expiry job to expire snapshots older than 5-7 days, given that the compaction job runs daily. This eliminates data duplication and reduces storage costs.
We've built a PySpark script that automates this entire process. Here's how it works:
Workflow Overview
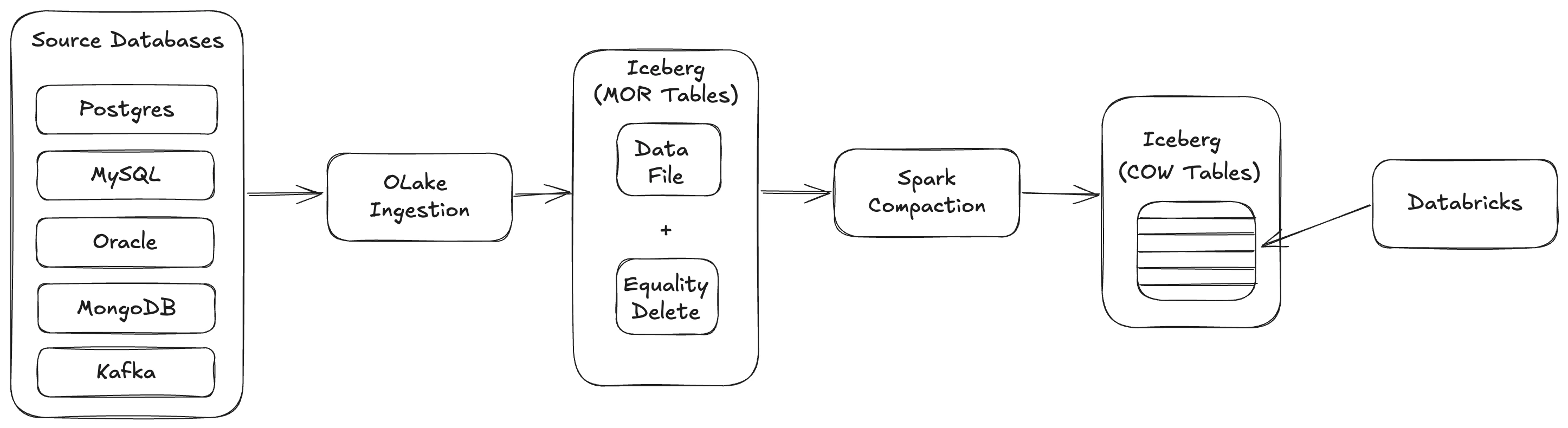
The workflow consists of the following steps:
- Data Ingestion: Multiple source databases (PostgreSQL, MySQL, Oracle, MongoDB, Kafka) are ingested through OLake
- MOR Table Creation: OLake creates MOR-Equality-delete-tables
- COW Write: Spark script to transforms MOR tables into Copy-on-Write (COW) format by rewriting data files with equality deletes applied
- Storage: COW tables are stored in object storage (S3, Azure Blob Storage, GCS, etc.)
- Querying: Databricks queries COW tables as external Iceberg tables with all deletes and updates properly applied
Prerequisites
Before running the MOR to COW write script, ensure you have the following installed:
- Java 21: Required for Spark runtime
- Python 3.13.7: Required for PySpark
- Spark 3.5.2: Apache Spark with Iceberg support
Additionally, make sure you have:
- Permission to access the Iceberg catalog
- Access to the object storage (S3, Azure Blob Storage, GCS, etc.) where your Iceberg tables are stored
- Appropriate cloud provider credentials or IAM roles configured
Generate Destination Details
Before running the MOR to COW write script, you need to generate a destination.json file that contains your catalog configuration and credentials. This file is required as input for the write script.
View the destination details generation script
#!/bin/bash
# ============================================================================
# CONFIGURATION: Edit this section to customize the script
# ============================================================================
# API endpoint base URL
# Example: BASE_URL="http://localhost:8000"
# BASE_URL="http://api.example.com"
BASE_URL="http://localhost:8000"
# OLake credentials
# Example: USERNAME="admin"
# PASSWORD="your_password"
USERNAME="<YOUR_USERNAME>"
PASSWORD="<YOUR_PASSWORD>"
# Job ID to query (can also be provided as command line argument)
# Example: JOB_ID=157
# Usage: ./get_destination_destination.sh [job_id]
# If job_id is provided as argument, it overrides this value
JOB_ID=9
# ============================================================================
# Check if jq is available
if ! command -v jq &> /dev/null; then
echo "Error: jq is required but not installed. Please install jq to use this script."
exit 1
fi
# Get job ID from command line argument if provided, otherwise use script variable
if [ -n "$1" ]; then
JOB_ID="$1"
fi
# Check if job ID is specified
if [ -z "$JOB_ID" ] || [ "$JOB_ID" == "" ]; then
echo "Error: Please specify a job ID either in the script (JOB_ID variable) or as a command line argument."
echo "Usage: $0 [job_id]"
exit 1
fi
# Login and save cookies
echo "Logging in to $BASE_URL..."
curl --location "$BASE_URL/login" \
--header 'Content-Type: application/json' \
--data "{
\"username\": \"$USERNAME\",
\"password\": \"$PASSWORD\"
}" \
-c cookies.txt \
-s > /dev/null
# Get jobs data and save to temporary file
echo "Fetching jobs data for job ID: $JOB_ID..."
RESPONSE_FILE=$(mktemp)
curl --location "$BASE_URL/api/v1/project/123/jobs" \
--header 'Content-Type: application/json' \
-b cookies.txt \
-s > "$RESPONSE_FILE"
# Extract and save destination.config as parsed JSON (single object, not array)
OUTPUT_FILE="destination.json"
jq -r ".data[]? | select(.id == $JOB_ID) |
.destination.config // \"\" |
if type == \"string\" and length > 0 then
fromjson
else
{}
end" "$RESPONSE_FILE" 2>/dev/null > "$OUTPUT_FILE"
echo "Results saved to: $OUTPUT_FILE"
# Cleanup
rm -f "$RESPONSE_FILE" cookies.txt
Before running the script, update the following configuration:
BASE_URL: Replace with your OLake API endpoint URL (e.g.,http://localhost:8000or your production API URL)USERNAME: Replace<YOUR_USERNAME>with your OLake usernamePASSWORD: Replace<YOUR_PASSWORD>with your OLake passwordJOB_ID: Replace with your actual job ID. The job ID can be obtained from the OLake UI job section. This job ID is required to retrieve the catalog configuration, URIs, and credentials fromdestination.jsonfor all tables synced by that job.
To run the script:
- Save the script to a file (e.g.,
get_destination_details.sh) in your desired directory - Navigate to the directory where you saved the script:
cd /path/to/your/script/directory - Make the script executable (if needed):
chmod +x get_destination_details.sh - Run the script:
Or if you made it executable:
bash get_destination_details.shYou can also pass the job ID as a command line argument:./get_destination_details.sh./get_destination_details.sh <job_id>
The script will generate a destination.json file that contains the catalog configuration, credentials, and object storage settings needed by the MOR to COW write script. This file is automatically used by the write script to configure the Spark session with the correct Iceberg catalog and storage credentials.
MOR to COW Write Script
📥 Download mor_to_cow_script.pyView the PySpark MOR to COW write script
import argparse
import json
import os
from typing import Optional, Tuple, Union, List
from pyspark.sql import SparkSession
from pyspark.sql.utils import AnalysisException
# Spark session is created in __main__ after we parse destination config.
spark = None # type: ignore[assignment]
# ------------------------------------------------------------------------------
# User Inputs (must be provided)
# ------------------------------------------------------------------------------
CATALOG = "olake_iceberg"
# Source namespace/database for MOR tables.
# User must hardcode this before running the script.
DB = "<NAME_OF_YOUR_SOURCE_DATABASE>"
# Destination namespace/database for generated COW tables (same catalog, different db)
COW_DB = "<NAME_OF_YOUR_COW_DATABASE>"
# Base S3 location where per-table COW tables (and the shared state table) will be stored.
# Example: "s3://my-bucket/warehouse/cow"
COW_BASE_LOCATION = "<YOUR_COW_BASE_LOCATION>"
# We use WAP (Write-Audit-Publish) pattern to store checkpoint state.
# The truncate snapshot_id is stored as the WAP ID, which is published after each successful write.
PRIMARY_KEY = "_olake_id"
def _recompute_derived_names():
# No derived names needed for state-table anymore.
return
def load_destination_writer_config(destination_details_path: str) -> dict:
with open(destination_details_path, "r", encoding="utf-8") as f:
outer = json.load(f)
if not isinstance(outer, dict):
raise ValueError("Destination config file must be a JSON object with a 'writer' object")
writer = outer.get("writer")
if not isinstance(writer, dict):
raise ValueError("Destination config JSON does not contain a 'writer' object")
return writer
def _normalize_warehouse(catalog_type: str, warehouse_val: str) -> str:
"""
- REST/Lakekeeper: warehouse can be a Lakekeeper 'warehouse name' (not a URI).
- Glue/JDBC: warehouse must be a filesystem URI/path (often s3a://bucket/prefix).
"""
if not warehouse_val:
raise ValueError("iceberg_s3_path is required")
v = warehouse_val.strip()
if catalog_type == "rest":
return v
# For glue/jdbc, accept s3:// or s3a://; if no scheme, assume it's "bucket/prefix"
if v.startswith("s3://"):
return "s3a://" + v[len("s3://") :]
if v.startswith("s3a://"):
return v
return "s3a://" + v.lstrip("/")
def _spark_packages_for(writer: dict, catalog_type: str) -> str:
"""
Base packages are required for Iceberg + S3. JDBC catalogs additionally need a DB driver.
"""
pkgs = [
"org.apache.iceberg:iceberg-spark-runtime-3.5_2.12:1.5.2",
"org.apache.iceberg:iceberg-aws-bundle:1.5.2",
"org.apache.hadoop:hadoop-aws:3.3.4",
"com.amazonaws:aws-java-sdk-bundle:1.12.262",
]
if catalog_type == "jdbc":
jdbc_url = (writer.get("jdbc_url") or "").lower()
# Common case for Iceberg JDBC catalog
if jdbc_url.startswith("jdbc:postgresql:"):
pkgs.append("org.postgresql:postgresql:42.5.4")
elif jdbc_url.startswith("jdbc:mysql:"):
pkgs.append("mysql:mysql-connector-java:8.0.33")
# de-dupe while preserving order
seen = set()
out = []
for p in pkgs:
if p not in seen:
seen.add(p)
out.append(p)
return ",".join(out)
def build_spark_session_from_writer(writer: dict) -> SparkSession:
catalog_type = (writer.get("catalog_type") or "").lower()
catalog_name = writer.get("catalog_name") or CATALOG
warehouse_raw = writer.get("iceberg_s3_path") or ""
warehouse = _normalize_warehouse(catalog_type, warehouse_raw)
# S3A settings
s3_endpoint = writer.get("s3_endpoint")
aws_region = writer.get("aws_region")
aws_access_key = writer.get("aws_access_key")
aws_secret_key = writer.get("aws_secret_key")
aws_session_token = (
writer.get("aws_session_token")
or writer.get("aws_sessionToken")
or writer.get("session_token")
or writer.get("sessionToken")
)
s3_path_style = writer.get("s3_path_style") # may not exist; we'll infer if missing
s3_use_ssl = writer.get("s3_use_ssl")
# Infer path-style for MinIO-like endpoints if not specified
if s3_path_style is None and isinstance(s3_endpoint, str):
if s3_endpoint.startswith("http://") or "9000" in s3_endpoint or "minio" in s3_endpoint.lower():
s3_path_style = True
if s3_path_style is None:
s3_path_style = True
# Infer SSL from endpoint scheme if present; allow explicit override via s3_use_ssl
ssl_enabled = None
if isinstance(s3_use_ssl, bool):
ssl_enabled = "true" if s3_use_ssl else "false"
if isinstance(s3_endpoint, str) and s3_endpoint.startswith("http://"):
ssl_enabled = ssl_enabled or "false"
elif isinstance(s3_endpoint, str) and s3_endpoint.startswith("https://"):
ssl_enabled = ssl_enabled or "true"
# Maven packages (network is available per your note)
packages = _spark_packages_for(writer, catalog_type)
builder = SparkSession.builder.appName("OLake MOR to COW Compaction")
builder = builder.config("spark.jars.packages", packages)
builder = builder.config(
"spark.sql.extensions",
"org.apache.iceberg.spark.extensions.IcebergSparkSessionExtensions",
)
builder = builder.config("spark.sql.catalogImplementation", "in-memory")
builder = builder.config("spark.sql.defaultCatalog", catalog_name)
# EMR/YARN resource management configs to prevent resource starvation
builder = builder.config("spark.dynamicAllocation.enabled", "true")
builder = builder.config("spark.dynamicAllocation.shuffleTracking.enabled", "true")
builder = builder.config("spark.dynamicAllocation.minExecutors", "1")
builder = builder.config("spark.dynamicAllocation.maxExecutors", "10")
builder = builder.config("spark.dynamicAllocation.initialExecutors", "2")
builder = builder.config("spark.dynamicAllocation.executorIdleTimeout", "60s")
builder = builder.config("spark.dynamicAllocation.schedulerBacklogTimeout", "1s")
builder = builder.config("spark.executor.instances", "2")
builder = builder.config("spark.executor.cores", "2")
builder = builder.config("spark.executor.memory", "2g")
builder = builder.config("spark.driver.memory", "2g")
builder = builder.config("spark.scheduler.maxRegisteredResourcesWaitingTime", "30s")
builder = builder.config("spark.scheduler.minRegisteredResourcesRatio", "0.5")
# Ensure AWS SDK-based clients (e.g., GlueCatalog, Iceberg S3FileIO) can see credentials.
# This avoids requiring users to export env vars in the container.
if aws_region:
os.environ["AWS_REGION"] = str(aws_region)
os.environ["AWS_DEFAULT_REGION"] = str(aws_region)
builder = builder.config("spark.driverEnv.AWS_REGION", str(aws_region))
builder = builder.config("spark.driverEnv.AWS_DEFAULT_REGION", str(aws_region))
builder = builder.config("spark.executorEnv.AWS_REGION", str(aws_region))
builder = builder.config("spark.executorEnv.AWS_DEFAULT_REGION", str(aws_region))
if aws_access_key and aws_secret_key:
os.environ["AWS_ACCESS_KEY_ID"] = str(aws_access_key)
os.environ["AWS_SECRET_ACCESS_KEY"] = str(aws_secret_key)
builder = builder.config("spark.driverEnv.AWS_ACCESS_KEY_ID", str(aws_access_key))
builder = builder.config("spark.driverEnv.AWS_SECRET_ACCESS_KEY", str(aws_secret_key))
builder = builder.config("spark.executorEnv.AWS_ACCESS_KEY_ID", str(aws_access_key))
builder = builder.config("spark.executorEnv.AWS_SECRET_ACCESS_KEY", str(aws_secret_key))
if aws_session_token:
os.environ["AWS_SESSION_TOKEN"] = str(aws_session_token)
builder = builder.config("spark.driverEnv.AWS_SESSION_TOKEN", str(aws_session_token))
builder = builder.config("spark.executorEnv.AWS_SESSION_TOKEN", str(aws_session_token))
# SparkCatalog wrapper
builder = builder.config(f"spark.sql.catalog.{catalog_name}", "org.apache.iceberg.spark.SparkCatalog")
builder = builder.config(f"spark.sql.catalog.{catalog_name}.io-impl", "org.apache.iceberg.aws.s3.S3FileIO")
builder = builder.config(f"spark.sql.catalog.{catalog_name}.warehouse", warehouse)
# IMPORTANT: Iceberg's S3FileIO uses AWS SDK directly (not Hadoop S3A configs).
# For MinIO/non-AWS endpoints, set Iceberg catalog-level s3.* properties so
# metadata/data writes go to the correct endpoint.
builder = builder.config(
f"spark.sql.catalog.{catalog_name}.s3.path-style-access",
str(bool(s3_path_style)).lower(),
)
if s3_endpoint:
builder = builder.config(f"spark.sql.catalog.{catalog_name}.s3.endpoint", s3_endpoint)
if aws_region:
builder = builder.config(f"spark.sql.catalog.{catalog_name}.s3.region", aws_region)
if aws_access_key and aws_secret_key:
builder = builder.config(f"spark.sql.catalog.{catalog_name}.s3.access-key-id", aws_access_key)
builder = builder.config(f"spark.sql.catalog.{catalog_name}.s3.secret-access-key", aws_secret_key)
# Catalog impl specifics
if catalog_type == "rest":
rest_url = writer.get("rest_catalog_url")
if not rest_url:
raise ValueError("rest_catalog_url is required for catalog_type=rest")
builder = builder.config(f"spark.sql.catalog.{catalog_name}.catalog-impl", "org.apache.iceberg.rest.RESTCatalog")
builder = builder.config(f"spark.sql.catalog.{catalog_name}.uri", rest_url)
elif catalog_type == "glue":
builder = builder.config(
f"spark.sql.catalog.{catalog_name}.catalog-impl",
"org.apache.iceberg.aws.glue.GlueCatalog",
)
# Optional: Glue catalog id/account id if provided
glue_catalog_id = writer.get("glue_catalog_id") or writer.get("glue.catalog-id") or writer.get("catalog_id")
if glue_catalog_id:
builder = builder.config(f"spark.sql.catalog.{catalog_name}.glue.catalog-id", str(glue_catalog_id))
# Region can be needed by AWS SDK for Glue
if aws_region:
builder = builder.config(
"spark.driver.extraJavaOptions",
f"-Daws.region={aws_region} -Daws.defaultRegion={aws_region}",
)
builder = builder.config(
"spark.executor.extraJavaOptions",
f"-Daws.region={aws_region} -Daws.defaultRegion={aws_region}",
)
elif catalog_type == "jdbc":
jdbc_url = writer.get("jdbc_url")
if not jdbc_url:
raise ValueError("jdbc_url is required for catalog_type=jdbc")
builder = builder.config(
f"spark.sql.catalog.{catalog_name}.catalog-impl",
"org.apache.iceberg.jdbc.JdbcCatalog",
)
builder = builder.config(f"spark.sql.catalog.{catalog_name}.uri", jdbc_url)
jdbc_user = writer.get("jdbc_username") or writer.get("jdbc_user") or writer.get("username")
jdbc_password = writer.get("jdbc_password") or writer.get("jdbc_pass") or writer.get("password")
if jdbc_user:
builder = builder.config(f"spark.sql.catalog.{catalog_name}.jdbc.user", str(jdbc_user))
if jdbc_password:
builder = builder.config(f"spark.sql.catalog.{catalog_name}.jdbc.password", str(jdbc_password))
else:
raise ValueError(f"Unsupported catalog_type={catalog_type}. Supported: rest, glue, jdbc")
# S3A filesystem settings
builder = builder.config("spark.hadoop.fs.s3a.impl", "org.apache.hadoop.fs.s3a.S3AFileSystem")
builder = builder.config("spark.hadoop.fs.s3a.path.style.access", str(bool(s3_path_style)).lower())
if s3_endpoint:
builder = builder.config("spark.hadoop.fs.s3a.endpoint", s3_endpoint)
if aws_region:
builder = builder.config("spark.hadoop.fs.s3a.region", aws_region)
if ssl_enabled is not None:
builder = builder.config("spark.hadoop.fs.s3a.connection.ssl.enabled", ssl_enabled)
if aws_access_key and aws_secret_key:
builder = builder.config(
"spark.hadoop.fs.s3a.aws.credentials.provider",
"org.apache.hadoop.fs.s3a.SimpleAWSCredentialsProvider",
)
builder = builder.config("spark.hadoop.fs.s3a.access.key", aws_access_key)
builder = builder.config("spark.hadoop.fs.s3a.secret.key", aws_secret_key)
return builder.getOrCreate()
# ------------------------------------------------------------------------------
# Helpers
# ------------------------------------------------------------------------------
def split_fqn(table_fqn: str):
parts = table_fqn.split(".")
if len(parts) != 3:
raise ValueError(f"Expected table fqn as <catalog>.<db>.<table>, got: {table_fqn}")
return parts[0], parts[1], parts[2]
def cow_table_and_location_for(mor_table_fqn: str):
catalog, _db, table = split_fqn(mor_table_fqn)
cow_table_fqn = f"{catalog}.{COW_DB}.{table}_cow"
cow_location = f"{COW_BASE_LOCATION}/{table}_cow"
return cow_table_fqn, cow_location
def table_exists(table_name: str) -> bool:
try:
spark.read.format("iceberg").load(table_name).limit(1).collect()
return True
except AnalysisException:
return False
def ensure_namespace_exists(catalog: str, namespace: str):
# Create destination namespace for COW tables/state if missing.
# Iceberg SparkCatalog supports CREATE NAMESPACE for REST/Glue catalogs.
spark.sql(f"CREATE NAMESPACE IF NOT EXISTS {catalog}.{namespace}")
def enable_wap_for_table(cow_table_fqn: str):
"""Enable WAP (Write-Audit-Publish) for the COW table if not already enabled."""
try:
spark.sql(f"ALTER TABLE {cow_table_fqn} SET TBLPROPERTIES ('write.wap.enabled'='true')")
except Exception:
# WAP might already be enabled, ignore error
pass
def get_wap_id_from_table(cow_table_fqn: str, catalog_name: str) -> Optional[str]:
"""
Get the latest published WAP ID from the COW table.
Returns the WAP ID (string) or None if no WAP ID exists.
Queries the COW table's snapshot metadata to find WAP IDs stored in snapshot summaries.
"""
if not table_exists(cow_table_fqn):
return None
# Check snapshot metadata for WAP ID
try:
rows = spark.sql(f"""
SELECT summary
FROM {cow_table_fqn}.snapshots
WHERE summary IS NOT NULL
ORDER BY committed_at DESC
LIMIT 10
""").collect()
for r in rows:
d = r.asDict(recursive=True)
summary = d.get("summary") or {}
if isinstance(summary, dict):
# Look for wap_id in snapshot summary (check multiple key variations)
wap_id = summary.get("wap.id") or summary.get("wap_id") or summary.get("wap-id")
if wap_id:
return str(wap_id)
except Exception:
pass
return None
def publish_wap_changes(cow_table_fqn: str, catalog_name: str, wap_id: str):
"""
Publish WAP changes. Idempotent - can be called multiple times safely.
Catches duplicate WAP commit errors and cherry-pick validation errors (occurs when re-publishing already published WAP IDs).
"""
try:
spark.sql(f"CALL {catalog_name}.system.publish_changes('{cow_table_fqn}', '{wap_id}')")
except Exception as e:
error_msg = str(e).lower()
# DuplicateWAPCommitException: "Duplicate request to cherry pick wap id that was published already"
# Cherry-pick validation errors: "cannot cherry-pick" or "not append, dynamic overwrite, or fast-forward"
# Both indicate the WAP ID is already published, which is idempotent
if ("duplicate" in error_msg and "wap" in error_msg and "published already" in error_msg) or \
"cannot cherry-pick" in error_msg or \
"not append, dynamic overwrite, or fast-forward" in error_msg:
# Idempotent - already published, that's fine
print(f"[{cow_table_fqn}] WAP ID {wap_id} already published (idempotent operation).")
else:
# Re-raise if it's a different error
raise
def extract_truncate_id_from_wap_id(wap_id: str) -> Optional[int]:
"""Extract truncate snapshot_id from WAP ID. WAP ID should be the truncate snapshot_id itself."""
try:
return int(wap_id)
except Exception:
return None
# ------------------------------------------------------------------------------
# Iceberg snapshot helpers
# ------------------------------------------------------------------------------
def get_latest_snapshot_and_parent_id(table_fqn: str):
"""
Return the most recent TRUNCATE-like snapshot (snapshot_id, parent_id).
To be robust against a small race where new OLake writes are committed
immediately after our TRUNCATE, we look at the latest few snapshots and
pick the first one that matches the truncate boundary signature.
"""
rows = spark.sql(f"""
SELECT snapshot_id, parent_id, committed_at, operation, summary
FROM {table_fqn}.snapshots
ORDER BY committed_at DESC
LIMIT 10
""").collect()
if not rows:
return None, None
snaps = []
by_id = {}
for r in rows:
d = r.asDict(recursive=True)
snap = {
"snapshot_id": d.get("snapshot_id"),
"parent_id": d.get("parent_id"),
"committed_at": d.get("committed_at"),
"operation": d.get("operation"),
"summary": d.get("summary"),
}
snaps.append(snap)
sid = snap.get("snapshot_id")
if sid is not None:
by_id[sid] = snap
# Among these most recent snapshots, find the newest one that looks like a truncate.
for snap in snaps:
parent = by_id.get(snap.get("parent_id"))
if _is_truncate_boundary_snapshot(snap, parent):
return snap.get("snapshot_id"), snap.get("parent_id")
# Fallback: if none looks like a truncate, just return the latest snapshot.
head = snaps[0]
return head.get("snapshot_id"), head.get("parent_id")
def _summary_int(summary: Optional[dict], key: str) -> Optional[int]:
if not summary or key not in summary:
return None
try:
return int(summary.get(key)) # type: ignore[arg-type]
except Exception:
return None
def _summary_first_int(summary: Optional[dict], keys: Tuple[str, ...]) -> Optional[int]:
if not summary:
return None
for k in keys:
v = _summary_int(summary, k)
if v is not None:
return v
return None
def _added_delete_files(summary: Optional[dict]) -> int:
"""
Best-effort: different engines/versions may emit different keys.
Treat missing keys as 0.
"""
if not summary:
return 0
for k in ("added-delete-files", "added-equality-delete-files", "added-position-delete-files"):
v = _summary_int(summary, k)
if v is not None and v != 0:
return v
# If keys exist but are '0', return 0.
return 0
def _removed_data_files(summary: Optional[dict]) -> Optional[int]:
"""
Best-effort: removal count is sometimes stored as 'deleted-data-files' (Iceberg metrics),
and sometimes as other keys depending on engine.
"""
for k in ("deleted-data-files", "removed-data-files", "deleted_files", "removed_files"):
v = _summary_int(summary, k)
if v is not None:
return v
return None
def _removed_delete_files(summary: Optional[dict]) -> Optional[int]:
# Best-effort; key names vary by engine/version.
return _summary_first_int(
summary,
(
"deleted-delete-files",
"removed-delete-files",
"deleted_delete_files",
"removed_delete_files",
),
)
def _total_delete_files(summary: Optional[dict]) -> Optional[int]:
return _summary_first_int(summary, ("total-delete-files", "total_delete_files"))
def _is_truncate_boundary_snapshot(snap: dict, parent: Optional[dict]) -> bool:
"""
Identify compaction boundary snapshots created by TRUNCATE TABLE.
- operation in {'delete','overwrite'} (varies by engine/version)
- added-data-files == 0
- added delete files == 0
- total-data-files == 0 (table empty after boundary)
- removed/deleted data files == parent.total-data-files (when both are available)
- removed/deleted delete files == parent.total-delete-files (when both are available)
"""
op = (snap.get("operation") or "").lower()
if op not in ("delete", "overwrite"):
return False
summary = snap.get("summary") or {}
parent_summary = (parent or {}).get("summary") or {}
added_data_files = _summary_int(summary, "added-data-files") or 0
if added_data_files != 0:
return False
if _added_delete_files(summary) != 0:
return False
total_data_files = _summary_int(summary, "total-data-files")
if total_data_files is not None and total_data_files != 0:
return False
total_delete_files = _total_delete_files(summary)
if total_delete_files is not None and total_delete_files != 0:
return False
removed = _removed_data_files(summary)
parent_total = _summary_int(parent_summary, "total-data-files")
if removed is not None and parent_total is not None:
if removed != parent_total:
return False
removed_del = _removed_delete_files(summary)
parent_total_del = _total_delete_files(parent_summary)
if removed_del is not None and parent_total_del is not None:
if removed_del != parent_total_del:
return False
# Fallback if one side isn't available: delete-to-empty should remove something.
if removed is not None:
return removed > 0
# If the engine doesn't report removed data files, we can't reliably detect.
return False
# ------------------------------------------------------------------------------
# Merge + schema alignment
# ------------------------------------------------------------------------------
def align_cow_schema(cow_table_fqn: str, mor_df, cow_df):
mor_schema = {f.name: f.dataType for f in mor_df.schema.fields}
cow_schema = {f.name: f.dataType for f in cow_df.schema.fields}
for col, dtype in mor_schema.items():
if col not in cow_schema:
print(f"Adding new column '{col}' with type '{dtype.simpleString()}' to COW table")
spark.sql(f"""
ALTER TABLE {cow_table_fqn}
ADD COLUMN {col} {dtype.simpleString()}
""")
for col, mor_type in mor_schema.items():
if col in cow_schema:
cow_type = cow_schema[col]
if mor_type != cow_type:
print(
f"Updating column '{col}' type from '{cow_type.simpleString()}' "
f"to '{mor_type.simpleString()}' in COW table"
)
spark.sql(f"""
ALTER TABLE {cow_table_fqn}
ALTER COLUMN {col} TYPE {mor_type.simpleString()}
""")
def merge_snapshot_into_cow(mor_table_fqn: str, cow_table_fqn: str, snapshot_id: int):
mor_df = (
spark.read.format("iceberg")
.option("snapshot-id", snapshot_id)
.load(mor_table_fqn)
)
cow_df = spark.read.format("iceberg").load(cow_table_fqn)
align_cow_schema(cow_table_fqn, mor_df, cow_df)
spark.sql(f"""
MERGE INTO {cow_table_fqn} AS target
USING (
SELECT *
FROM {mor_table_fqn}
VERSION AS OF {snapshot_id}
) AS source
ON target.{PRIMARY_KEY} = source.{PRIMARY_KEY}
WHEN MATCHED THEN
UPDATE SET *
WHEN NOT MATCHED THEN
INSERT *
""")
def _cow_has_any_snapshots(cow_table_fqn: str) -> bool:
if not table_exists(cow_table_fqn):
return False
try:
cnt = spark.sql(f"SELECT COUNT(*) AS c FROM {cow_table_fqn}.snapshots").collect()[0]["c"]
return int(cnt) > 0
except Exception:
# If metadata table isn't accessible for some reason, assume it has snapshots.
return True
def _fetch_snapshot_with_summary(table_fqn: str, snapshot_id: int) -> Optional[dict]:
"""
Fetch a single snapshot (and its summary) by snapshot_id.
"""
rows = spark.sql(f"""
SELECT snapshot_id, parent_id, committed_at, operation, summary
FROM {table_fqn}.snapshots
WHERE snapshot_id = {snapshot_id}
""").collect()
if not rows:
return None
d = rows[0].asDict(recursive=True)
return {
"snapshot_id": d.get("snapshot_id"),
"parent_id": d.get("parent_id"),
"committed_at": d.get("committed_at"),
"operation": d.get("operation"),
"summary": d.get("summary"),
}
def _set_wap_id(wap_id: Optional[Union[int, str]]):
if wap_id is None:
spark.sql("SET spark.wap.id=")
else:
spark.sql(f"SET spark.wap.id={wap_id}")
def _ensure_cow_table_from_snapshot(
mor_table_fqn: str,
cow_table_fqn: str,
cow_location: str,
snapshot_id_for_schema: int,
):
"""
Create the COW table if missing by CTAS from a MOR snapshot.
This also anchors the initial schema for later schema alignment.
"""
if table_exists(cow_table_fqn):
enable_wap_for_table(cow_table_fqn)
return
spark.sql(f"""
CREATE TABLE {cow_table_fqn}
USING iceberg
LOCATION '{cow_location}'
TBLPROPERTIES ('write.wap.enabled'='true')
AS
SELECT *
FROM {mor_table_fqn}
VERSION AS OF {snapshot_id_for_schema}
""")
enable_wap_for_table(cow_table_fqn)
def _apply_truncate_boundary(
mor_table_fqn: str,
cow_table_fqn: str,
cow_location: str,
catalog_name: str,
boundary_snap: dict,
):
"""
For a truncate boundary snapshot t:
- Compact/merge its parent snapshot h into the COW table (unless h is also a boundary).
- Commit using WAP ID = t.snapshot_id, then publish (idempotent).
"""
t_id = boundary_snap.get("snapshot_id")
h_id = boundary_snap.get("parent_id")
print(f"[{mor_table_fqn}] Processing boundary {t_id}; parent(high-water)={h_id}")
if t_id is None:
return
if not table_exists(cow_table_fqn) or not _cow_has_any_snapshots(cow_table_fqn):
print(f"[{mor_table_fqn}] COW table missing/empty; creating baseline from snapshot {h_id} ...")
_set_wap_id(t_id)
_ensure_cow_table_from_snapshot(mor_table_fqn, cow_table_fqn, cow_location, int(h_id))
publish_wap_changes(cow_table_fqn, catalog_name, str(t_id))
_set_wap_id(None)
print(f"[{mor_table_fqn}] Published WAP changes with truncate {t_id}.")
return
print(f"[{mor_table_fqn}] Compacting snapshot {h_id} into existing COW ...")
_set_wap_id(t_id)
merge_snapshot_into_cow(mor_table_fqn, cow_table_fqn, int(h_id))
publish_wap_changes(cow_table_fqn, catalog_name, str(t_id))
_set_wap_id(None)
print(f"[{mor_table_fqn}] Published WAP changes with truncate {t_id}.")
def run_compaction_cycle_for_table(mor_table_fqn: str):
cow_table_fqn, cow_location = cow_table_and_location_for(mor_table_fqn)
catalog_name, _, _ = split_fqn(mor_table_fqn)
# Step 1: Resume checkpoint from COW's last WAP id; re-publish it (idempotent) to finalize any half-done runs.
if table_exists(cow_table_fqn):
enable_wap_for_table(cow_table_fqn)
wap_id = get_wap_id_from_table(cow_table_fqn, catalog_name)
last_success_t = None
if wap_id:
print(f"[{mor_table_fqn}] Found existing WAP ID: {wap_id}. Re-publishing (idempotent)...")
publish_wap_changes(cow_table_fqn, catalog_name, wap_id)
last_success_t = extract_truncate_id_from_wap_id(wap_id)
if last_success_t is not None:
print(f"[{mor_table_fqn}] Last successful truncate checkpoint: {last_success_t}")
else:
print(f"[{mor_table_fqn}] Warning: Could not parse WAP ID {wap_id} as truncate snapshot id. Starting from beginning.")
else:
print(f"[{mor_table_fqn}] No WAP ID found; starting from earliest MOR history.")
# Step 2/3: Truncate MOR to create the boundary for this run.
spark.sql(f"TRUNCATE TABLE {mor_table_fqn}")
head_snapshot_id, _ = get_latest_snapshot_and_parent_id(mor_table_fqn)
if head_snapshot_id is None:
print(f"[{mor_table_fqn}] No snapshots found; nothing to do.")
return
# Build lineage from the new truncate snapshot back to (but not including) last_success_t.
by_id: dict = {}
lineage: List[dict] = []
# Optionally fetch the checkpoint snapshot so we know its summary when detecting truncates.
if last_success_t is not None:
chk = _fetch_snapshot_with_summary(mor_table_fqn, int(last_success_t))
if chk is not None:
by_id[chk["snapshot_id"]] = chk
cur_id = head_snapshot_id
seen = set()
while cur_id is not None and cur_id not in seen:
seen.add(cur_id)
snap = _fetch_snapshot_with_summary(mor_table_fqn, int(cur_id))
if snap is None:
break
by_id[snap["snapshot_id"]] = snap
lineage.append(snap)
parent_id = snap.get("parent_id")
# Stop once we've reached the snapshot whose parent is the checkpoint; this ensures we only
# reprocess snapshots strictly after last_success_t.
if last_success_t is not None and parent_id == last_success_t:
break
cur_id = parent_id
if not lineage:
print(f"[{mor_table_fqn}] No snapshots to scan between checkpoint and current truncate; nothing to do.")
return
# Process in chronological order (oldest -> newest).
lineage.reverse()
any_boundary = False
for snap in lineage:
parent = by_id.get(snap.get("parent_id"))
is_boundary = _is_truncate_boundary_snapshot(snap, parent)
if not is_boundary:
continue
any_boundary = True
t_id = snap.get("snapshot_id")
_apply_truncate_boundary(
mor_table_fqn=mor_table_fqn,
cow_table_fqn=cow_table_fqn,
cow_location=cow_location,
catalog_name=catalog_name,
boundary_snap=snap,
)
if not any_boundary:
# If we couldn't detect any truncate boundaries by signature (summary keys missing/version diff),
# process the head snapshot once as a synthetic boundary so we compact its parent.
head_snap = by_id.get(head_snapshot_id)
if head_snap is None:
print(f"[{mor_table_fqn}] Head snapshot {head_snapshot_id} not found; nothing to do.")
return
print(
f"[{mor_table_fqn}] Warning: no truncate boundaries detected by signature; "
f"processing head snapshot {head_snapshot_id} once as boundary."
)
_apply_truncate_boundary(
mor_table_fqn=mor_table_fqn,
cow_table_fqn=cow_table_fqn,
cow_location=cow_location,
catalog_name=catalog_name,
boundary_snap=head_snap,
)
def list_tables_in_db(catalog: str, db: str):
rows = spark.sql(f"SHOW TABLES IN {catalog}.{db}").collect()
table_names = []
for r in rows:
d = r.asDict(recursive=True)
if d.get("isTemporary", False):
continue
name = d.get("tableName") or d.get("table")
if name:
table_names.append(name)
return table_names
# ------------------------------------------------------------------------------
# Entry Point
# ------------------------------------------------------------------------------
if __name__ == "__main__":
parser = argparse.ArgumentParser(description="MOR -> COW compaction (REST Lakekeeper / Glue), configured from destination_details.json")
parser.add_argument(
"--destination-details",
required=True,
help="Path to destination_details.json generated by get_destination_details.sh",
)
parser.add_argument("--cow-db", default=COW_DB, help="Destination namespace/database for COW tables/state")
parser.add_argument("--catalog-name", default=None, help="Override catalog name (otherwise taken from destination config)")
args = parser.parse_args()
# Source DB is expected to be hardcoded in this file.
if not DB or DB.strip() == "" or DB.strip() == "<YOUR_SOURCE_DB>":
raise ValueError("Please set DB = '<YOUR_SOURCE_DB>' at the top of fail_test.py before running.")
# Update globals from args
COW_DB = args.cow_db
writer = load_destination_writer_config(args.destination_details)
if args.catalog_name:
writer["catalog_name"] = args.catalog_name
# Update catalog name global (used in derived FQNs)
CATALOG = writer.get("catalog_name") or CATALOG
_recompute_derived_names()
# Create Spark session with the right Iceberg/S3 config
spark = build_spark_session_from_writer(writer)
# Ensure destination namespace exists before creating state/COW tables
ensure_namespace_exists(CATALOG, COW_DB)
# Always compact all MOR tables in the source namespace/database.
all_tables = list_tables_in_db(CATALOG, DB)
mor_tables = [
f"{CATALOG}.{DB}.{t}"
for t in all_tables
if not t.endswith("_cow")
]
successes = []
failures = []
for mor_table in mor_tables:
try:
run_compaction_cycle_for_table(mor_table)
successes.append(mor_table)
except Exception as e:
failures.append((mor_table, str(e)))
print(f"[{mor_table}] FAILED: {e}")
print("---- Compaction Summary ----")
print(f"Successful tables: {len(successes)}")
for t in successes:
print(f" - {t}")
print(f"Failed tables: {len(failures)}")
for t, err in failures:
print(f" - {t}: {err}")
Before running the script, make sure to update the following variables in the "User Inputs" section:
CATALOG: Replaceolake_icebergwith the name of the catalog where the MOR tables live (e.g.,olake_iceberg)COW_DB: Replace<NAME_OF_YOUR_COW_DATABASE>with the namespace/database where COW tables + the state table should live (e.g.,postgres_main_public_cow)COW_BASE_LOCATION: Replace<YOUR_COW_BASE_LOCATION>with the base object-storage path where COW tables will be written (e.g.,s3://my-bucket/warehouse/cow)DB: Replace<NAME_OF_YOUR_SOURCE_DATABASE>with the namespace/database containing the MOR tables to be converted to COW format
For Lakekeeper catalogs, COW_BASE_LOCATION must be within the warehouse location. You can use a subfolder (e.g., if warehouse is s3://bucket/warehouse, use s3://bucket/warehouse/cow).
To execute the MOR to COW write script, use the following spark-submit command:
spark-submit \
--master 'local[*]' \
compaction_script.py \
--destination-details destination.json
Replace compaction_script.py with the actual name of your MOR to COW write script file. The script will automatically read the catalog configuration, credentials, and object storage settings from the destination.json file generated by the previous step.
If you're running the script on AWS EMR, you need to make sure:
-
Copy files to S3: Copy both
destination.jsonand the MOR to COW write script (e.g.,mor_to_cow_script.py) to an S3 bucket that your EMR cluster can access. -
Include EMR-specific Spark configurations: When submitting the job, include the following Spark configurations to prevent resource starvation and ensure optimal performance:
builder = builder.config("spark.dynamicAllocation.enabled", "true")
builder = builder.config("spark.dynamicAllocation.shuffleTracking.enabled", "true")
builder = builder.config("spark.dynamicAllocation.minExecutors", "1")
builder = builder.config("spark.dynamicAllocation.maxExecutors", "10")
builder = builder.config("spark.dynamicAllocation.initialExecutors", "2")
builder = builder.config("spark.dynamicAllocation.executorIdleTimeout", "60s")
builder = builder.config("spark.dynamicAllocation.schedulerBacklogTimeout", "1s")
builder = builder.config("spark.executor.instances", "2")
builder = builder.config("spark.executor.cores", "2")
builder = builder.config("spark.executor.memory", "2g")
builder = builder.config("spark.driver.memory", "2g")
builder = builder.config("spark.scheduler.maxRegisteredResourcesWaitingTime", "30s")
builder = builder.config("spark.scheduler.minRegisteredResourcesRatio", "0.5")
These configurations ensure proper resource management and prevent job failures due to resource starvation on EMR clusters.
How the MOR to COW Write Script Works
The MOR to COW write process is designed to be safe, repeatable, and compatible with continuous CDC ingestion. Key features:
- Non-intrusive: Works alongside OLake's ongoing syncs using Iceberg's snapshot isolation
- Unified incremental mode: Uses a single function that handles both first-time and subsequent runs. On the first run, it checks if the COW table exists—if not, it creates the COW table with a full resolved dataset from the MOR table. On subsequent runs, it updates the existing COW table with only the latest changes.
1. Read the last successful truncate ID from the COW table: The process starts by checking the COW table metadata to determine whether a previous MOR → COW run has completed successfully.
- The COW table's snapshot history is scanned to find the most recent WAP ID stored in the snapshot summary (under the key wap.id).
- The WAP ID contains the truncate snapshot ID from the MOR table that was successfully processed and published.
- If no COW table exists, or if the table exists but no WAP ID is found in any snapshot, the system treats this as a first-time run.
- This step defines from where the next processing should begin.
2. Re-publish the last WAP ID: If a WAP ID was found in Step 1, the script re-publishes it using Iceberg's publish_changes procedure.
- This operation is idempotent—if the WAP ID is already published, Iceberg recognizes it and continues without error.
- This ensures that in any case if WAP ID was not published in the previous run even after data been written to COW table, it will be published in the current run.
- After re-publishing, the truncate snapshot ID is extracted from the WAP ID to determine the starting point for the current run.
3. Decide the starting snapshot in the MOR table: Based on the outcome of Steps 1 and Step 2:
- First Run (no COW table exists / no WAP ID found), the script starts from the earliest snapshot in the MOR table.
- Subsequent Run (COW table exists / WAP ID found), the script starts from the snapshot after the last successfully published truncate id.
This ensures the process never reprocesses already-handled data.
4. Truncate the MOR table to mark the current boundary: Before any data is merged, the MOR table is explicitly truncated.
- This truncate operation creates a new snapshot, which serves as the upper boundary for the current processing cycle.
- At this point, the workflow has a starting point (from Step 3) and an ending point (this newly created truncate).
5. Iterate through MOR snapshots and detect truncate boundaries: The workflow now walks through MOR snapshots sequentially, starting from the snapshot chosen in Step 3 and stopping at the truncate snapshot created in Step 4. During this iteration:
- Each snapshot is examined to determine whether it represents a truncate operation.
- Truncate snapshots are detected using metadata signals such as:
operation=deleteadded-data-files= 0added-delete-files= 0total-data-files= 0removed-data-files=previous_snapshot.total-data-filesThis allows the workflow to correctly identify all truncate boundaries, including those created in previous failed runs.
6. Prepare COW database, table, and schema: Before any data transfer happens, the workflow prepares the COW side completely.
- If this is the first run, the COW database is created if it does not exist.
- If the COW table already exists, the MOR schema is compared with the COW schema. If there are any schema changes, the COW table is altered to match the MOR table schema (new columns are added, column types are updated as needed).
This step guarantees that the COW table is fully ready and schema-compatible before any transfer begins.
7. Merge MOR data into the COW table with atomic checkpointing: Once the COW table and schema are prepared, the workflow transfers data for each detected truncate boundary using Iceberg's WAP pattern for atomic checkpointing.
For each truncate boundary in the processing range:
- Check for redundant boundaries: If a truncate snapshot's parent snapshot is also a truncate, it is skipped to avoid redundant processing.
- Set the WAP ID: Before writing data, the script sets
spark.wap.idto the current truncate snapshot ID. This ensures the upcoming write operation will be staged under this WAP ID. - Transfer data:
- First Time Transfer: Creates a new COW table and writes the initial data from the MOR table. The WAP ID (truncate snapshot ID) is appended to the metadata in a single commit to the COW table.
- Subsequent Transfer: Merges records from the MOR table into the existing COW table and updates the WAP ID with the current truncate snapshot ID for which the MOR to COW conversion is being performed.
- Publish the WAP changes: After the data write completes, the script calls Iceberg's
publish_changesprocedure with the truncate snapshot ID as the WAP ID. - Unset the WAP ID: The script clears
spark.wap.idto prepare for the next boundary.
Failure Recovery and State Management
View failure recovery and state management details
The MOR to COW write flow is designed to be safe to re-run by checkpointing progress only after a truncate cycle is fully and successfully transferred.
The workflow records successful truncate snapshot IDs in the data writes to COW table and stores them as WAP ID.
trunc0 → eq1 → eq2 → eq3 → eq4 → trunc1 → eq5 → eq6 → eq7 → trunc2
Here:
eq*snapshots contain cdc changestrunc*snapshots mark truncate boundaries
What the state table stores
For each MOR table its corresponding COW table stores:
**last_successful_truncate_snapshot_id**: The most recent truncate snapshot that has been fully merged into the COW table (e.g.,trunc1,trunc2)
On reruns, the script uses last_successful_truncate_snapshot_id as the effective checkpoint.
How recovery works
Let us assume that trunc0 was the most recent successful truncate operation and while running trunc1 the script failed. By the time we re run the script, OLake might have ingested some more CDC changes. This is how the workflow will behave:
- The script checks the COW table's snapshot history and finds the latest WAP ID containing
trunc0_snapshot_id. - It re-publishes this WAP ID to ensure the data written to COW will be visible to the query engine.
- Then it fetches the
trunc0stored in WAP ID and uses it as the starting point for the current run. The script truncates the MOR table again, creatingtrunc2as the boundary for the run. - It starts scanning the snapshots from
trunc0totrunc2finding for any truncate operations in this boundary. - While scanning it encounters
trunc1, it transfers the data from snapshot ofeq4to the COW table and also updates the WAP ID with the snapshot id oftrunc1. After this operation it publishes the changes to the COW table. - Then it continues its process to find next truncate operation and encounters
trunc2, it transfers the data from snapshot ofeq7to the COW table and also updates the WAP ID with the snapshot id oftrunc2. Similarly it publishes the changes to the COW table.
Running the MOR to COW Write Script
The MOR to COW write script is designed to run periodically, automatically keeping your COW table up-to-date with the latest changes from your MOR table. You can schedule this script as a cron job or through workflow orchestration tools, ensuring that your Databricks queries always reflect the most recent data according to your requirements.
Execution Platforms
The script can be run on any Spark cluster that has access to your Iceberg catalog and object storage (S3, Azure Blob Storage, GCS, etc.). Common execution platforms include:
- AWS EMR: Run the script as a Spark job on EMR clusters
- Databricks: Execute as a scheduled job in your Databricks workspace
- Local Spark: For testing or small-scale deployments
Simply submit the script using spark-submit with the appropriate Iceberg catalog configuration for your environment.
Scheduling the MOR to COW Write Job
The job execution frequency can be set based on your data freshness requirements and business needs. The script is idempotent, so you can run it as frequently as needed without worrying about duplicate processing. Here are some common scheduling patterns:
- Hourly: For real-time dashboards and analytics that require near-live data
- Every 6 hours: A balanced approach for most use cases, providing good data freshness without excessive compute costs
- Daily: Perfect for overnight batch reporting and scenarios where daily updates are sufficient
- On-demand: For low-volume tables or manual refresh workflows where you trigger the write job only when needed
You can configure the schedule using cron syntax, Airflow DAG schedules, or your preferred orchestration tool. Each run will process any new changes since the last write, keeping your COW table synchronized with your MOR table.
For Databricks users, once the COW table is created and being updated periodically, you simply create an external Iceberg table pointing to your COW table location, and you're ready to query with correct results—all deletes and updates properly applied.
Testing the MOR to COW Write Script Locally
View local testing steps
To understand how the MOR to COW write script works and see it in action, you can test it locally on your system before running it on production data. Follow these steps to run the script locally:
-
Use the following command to quickly spin up the source Postgres and destination (Iceberg/Parquet Writer) services using Docker Compose. This will download the required docker-compose files and start the containers in the background.
sh -c 'curl -fsSL https://raw.githubusercontent.com/datazip-inc/olake-docs/master/docs/community/docker-compose.yml -o docker-compose.source.yml && \
curl -fsSL https://raw.githubusercontent.com/datazip-inc/olake/master/destination/iceberg/local-test/docker-compose.yml -o docker-compose.destination.yml && \
docker compose -f docker-compose.source.yml --profile postgres -f docker-compose.destination.yml up -d'Once the containers are up and running, you can run the the below command to spin up the OLake UI:
curl -sSL https://raw.githubusercontent.com/datazip-inc/olake-ui/master/docker-compose.yml | docker compose -f - up -dNowt he OLake UI can be accessed at http://localhost:8000.
-
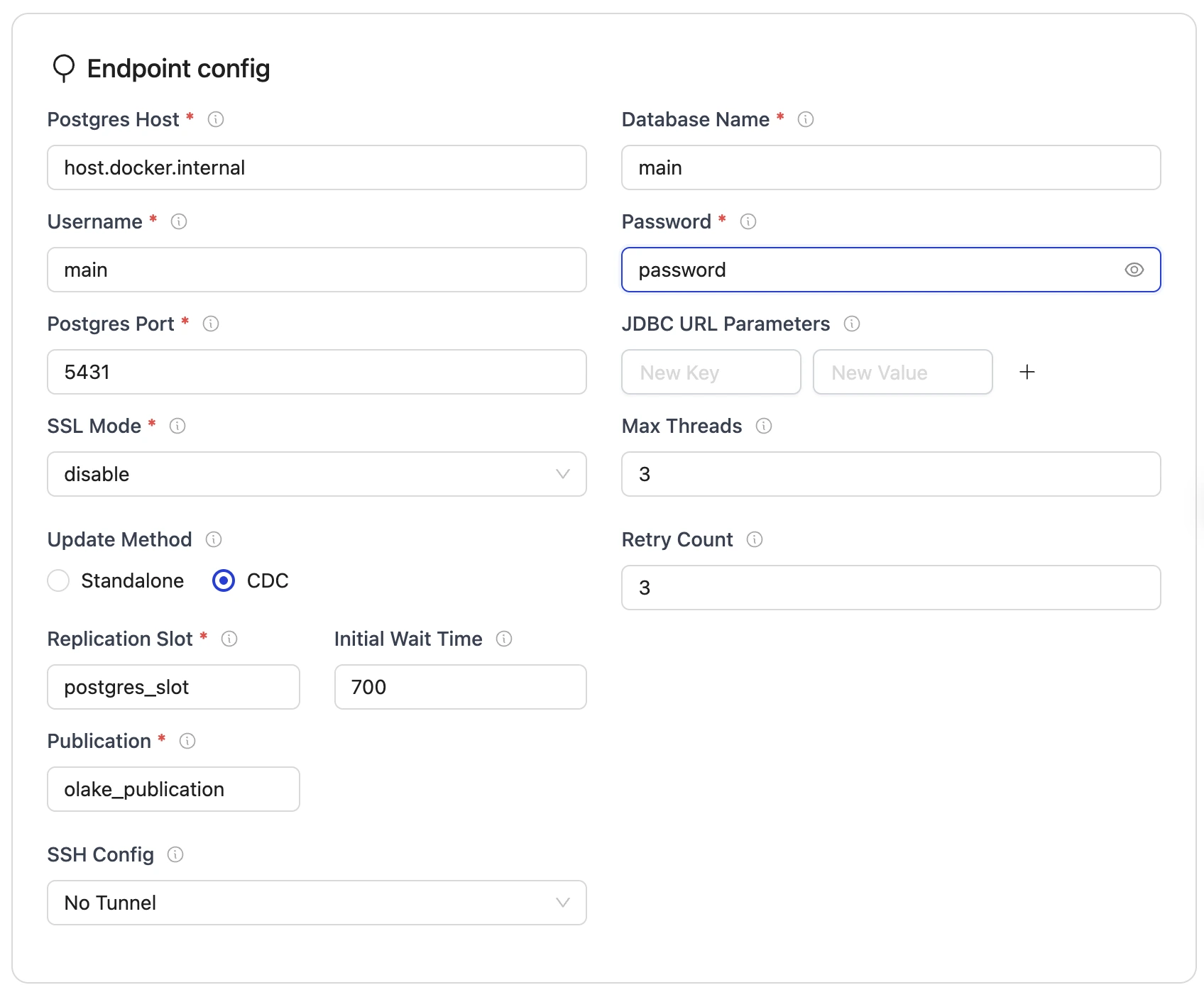
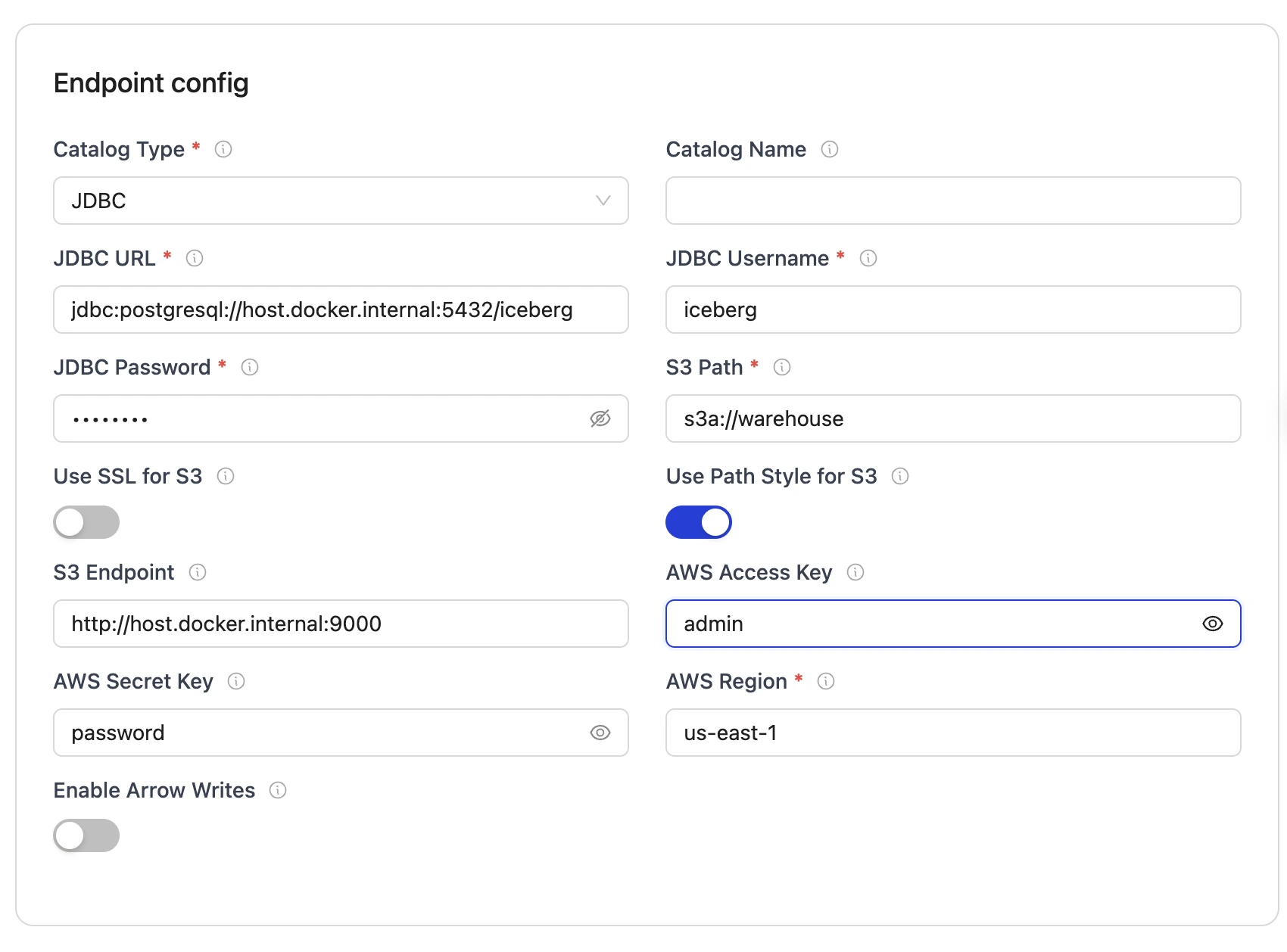
Set up the configuration:
- Source Configuration
- Destination Configuraiton
-
Select the streams and sync the data to Iceberg:
Since this is a local demo, we will sync the sample table
sample_datafrom the source database.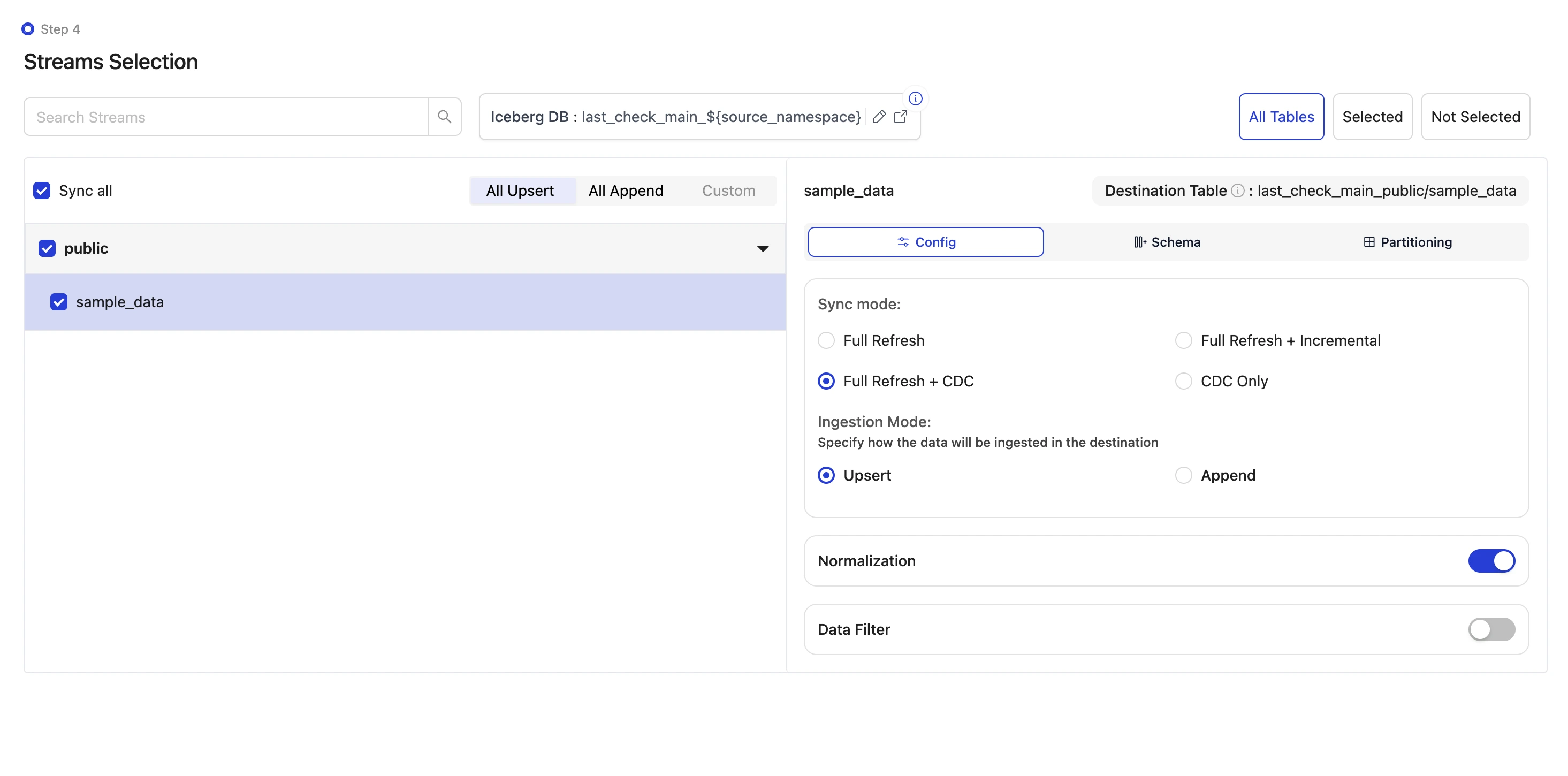
You can refer to the Streams Configuration for more details about the streams configuration and how to start the sync.
-
The data can be queried from Iceberg using the Spark Iceberg service available at
localhost:8888.To view the table run the following SQL command:
%%sql
SELECT * FROM olake_iceberg.postgres_main_public.sample_data;We can modify the source database by adding and modifying few records and then running the sync again with state enabled to see the changes in the Iceberg table.
Below command inserts two records into the source database:
docker exec -it primary_postgres psql -U main -d main -c "INSERT INTO public.sample_data (id, num_col, str_col) VALUES (10, 100, 'First record'), (20, 200, 'Second record');"Let us also update a record in the source database:
docker exec -it primary_postgres psql -U main -d main -c "UPDATE public.sample_data SET num_col = 150, str_col = 'First record updated' WHERE id = 1;"Now run the sync again. This can be done by simply clicking on the "Sync Now" button in the OLake UI.
To view the updated table run the following SQL command:
%%sql
SELECT * FROM olake_iceberg.postgres_main_public.sample_data; -
Run the MOR to COW write script to write COW tables:
After completing the historical load and CDC sync, your Iceberg table now contains both data files and equality delete files in object storage, representing a Merge-on-Read (MOR) table. To write this data as a Copy-on-Write (COW) table, run the MOR to COW write script with the following configuration:
Update the variables in the MOR to COW write script:
COW_DB = "postgres_main_public_cow"
COW_BASE_LOCATION = "s3a://warehouse/postgres_main_public_cow"Since we're running the script in the Spark Docker container, copy the required files to the container:
docker cp <PATH_TO_YOUR_WRITE_SCRIPT>/compaction_script.py spark-iceberg:/home/iceberg/compaction_script.py
docker cp <PATH_TO_YOUR_DESTINATION_DETAILS>/destination.json spark-iceberg:/home/iceberg/destination.jsonReplace
<PATH_TO_YOUR_WRITE_SCRIPT>and<PATH_TO_YOUR_DESTINATION_DETAILS>with the actual paths to your files on your local machine.Enter the Spark container:
docker exec -it spark-iceberg bashOnce inside the container, run the MOR to COW write script using spark-submit:
spark-submit \
--master 'local[*]' \
/home/iceberg/compaction_script.py \
--destination-details /home/iceberg/destination.json -
Verify the COW table creation:
Once the MOR to COW write script runs successfully, we can verify the results in MinIO (the local object storage used in this demo). It can be noticed that:
- The original MOR table with data files and equality delete files remains in
warehouse/postgres_main_public/sample_data - A new COW table has been created in
warehouse/postgres_main_public_cow/sample_data_cow, containing the resolved data with all equality deletes applied
You can verify the COW table by querying it in the Jupyter notebook available at
localhost:8888:%%sql
SELECT * FROM olake_iceberg.postgres_main_public_cow.sample_data_cow;This will display the resolved data from your new COW table with all equality deletes applied.
The COW table is now ready to be queried by Databricks as an external Iceberg table, with all updates and deletes properly reflected in the data files.
- The original MOR table with data files and equality delete files remains in
Conclusion
By implementing this automated MOR to COW write solution, you can now enjoy the best of both worlds: OLake's high-performance Merge-on-Read (MOR) writes for efficient CDC ingestion, combined with Databricks-compatible Copy-on-Write (COW) tables for accurate analytics queries.
