Aurora PostgreSQL CDC Setup Guide
Aurora PostgreSQL supports CDC through PostgreSQL logical decoding. In an Aurora cluster, CDC is enabled by turning on logical replication and using the native pgoutput plugin with a PostgreSQL publication to stream changes.
Prerequisites:
- An Amazon Aurora PostgreSQL cluster (PostgreSQL version 10+).
- Access to modify the cluster's parameter group (to enable logical replication).
- Ability to reboot the Aurora cluster (required for static parameter changes).
- The cluster should have Backups enabled (Aurora usually has continuous backups by default). This is important because binlog retention in Aurora is tied to backup retention.
- The RDS master user or another user with the
rds_superuserandrds_replicationroles.
Steps:
1. Enable Logical Replication in the Parameter Group
In the AWS RDS console, create or edit the cluster parameter group for your Aurora PostgreSQL cluster.
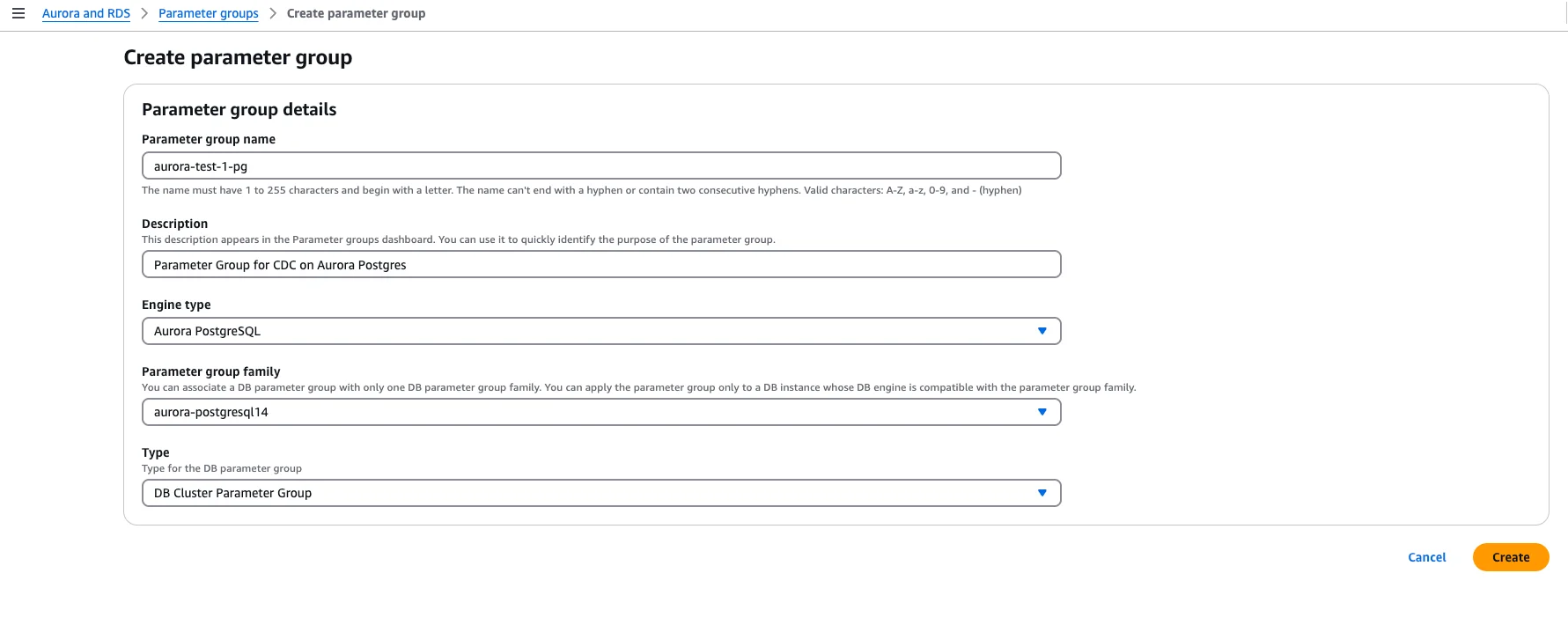
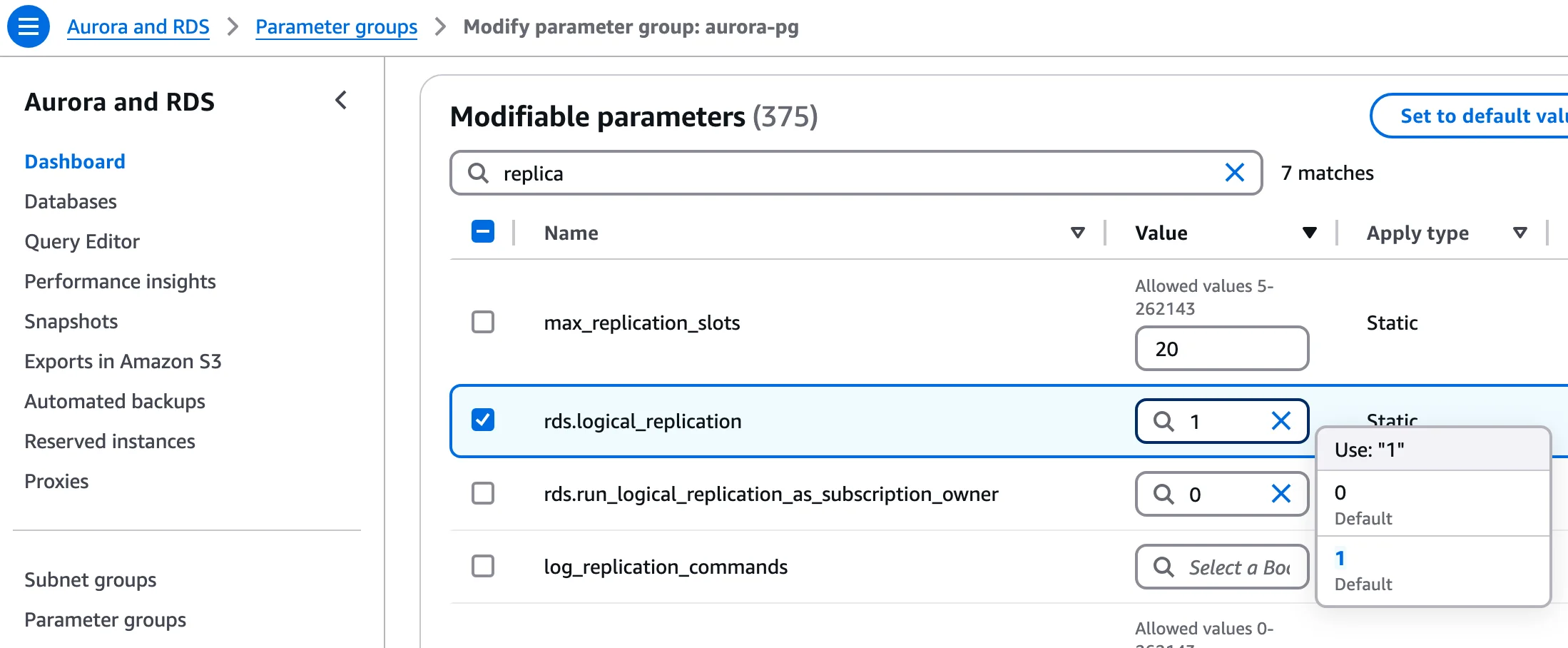
Configure these key parameters:
rds.logical_replication = 1- Enables logical replication (required for pgoutput)wal_level = logical- May be set automatically by the above parametermax_wal_sendersandmax_replication_slots- Set to accommodate your CDC connectionswal_sender_timeout = 0- (Optional) Prevents timeouts during long snapshots
Save the changes after configuration.
Turning on logical replication cannot be done on an Aurora read-replica instance. You must apply the change at the cluster level and then reboot the writer (primary) instance for the new parameter values to take effect. Restarting or modifying a reader instance alone will not enable logical decoding.
2. Reboot the Cluster to Apply Changes
rds.logical_replication is a static parameter, so reboot the Aurora cluster’s writer node to apply the new parameter group settings. After reboot, you can verify the setting by running:
SHOW rds.logical_replication;
It should return “on” (1)
3. Create or Grant a Replication-Privileged User
Use the master user (which has the rds_superuser role) or another user with sufficient privileges for CDC. Aurora’s default postgres user is an rds_superuser and has replication permissions by default. If you prefer to use a different user, create one and grant it the required roles:
CREATE USER cdc_user WITH PASSWORD 'strongpassword';
GRANT rds_superuser TO cdc_user;
-- (rds_superuser in Aurora automatically has logical replication permissions)
Aurora requires the CDC user to have superuser-level privileges. Use rds_superuser (Aurora's equivalent to SUPERUSER). Ensure network connectivity from your CDC client.
Optional: Read-only Privileges for Initial Snapshot
For stricter security, explicitly grant read access on schemas you plan to sync:
-- Example for the "public" schema in database mydb
GRANT CONNECT ON DATABASE mydb TO cdc_user;
GRANT USAGE ON SCHEMA public TO cdc_user;
GRANT SELECT ON ALL TABLES IN SCHEMA public TO cdc_user;
-- Ensure future tables are covered automatically
ALTER DEFAULT PRIVILEGES IN SCHEMA public GRANT SELECT ON TABLES TO cdc_user;
4. Create Publication
Publications define which tables will be replicated. Create a publication for the tables you want to monitor:
Publication Name Configuration:
Use the exact publication name that you configure in OLake UI or CLI. The publication name must match the publication parameter in your OLake source configuration.
-- Create publication for all tables in the database
CREATE PUBLICATION olake_publication FOR ALL TABLES;
-- Or create publication for specific tables
CREATE PUBLICATION olake_publication FOR TABLE public.table1, public.table2, public.table3;
-- Or create publication for all tables in a specific schema
CREATE PUBLICATION olake_publication FOR TABLES IN SCHEMA public;
Enable publish_via_partition_root for partitioned tables:
The publish_via_partition_root setting MUST be enabled to ensure data integrity when replicating partitioned tables. Without this setting, changes to partitioned tables will not be properly captured, resulting in data loss or inconsistency.
ALTER PUBLICATION olake_publication SET (publish_via_partition_root = true);
Publication Options:
-- Customize what operations are published
CREATE PUBLICATION olake_publication FOR ALL TABLES
WITH (publish = 'insert,update,delete,truncate');
-- Only publish INSERT and UPDATE operations
CREATE PUBLICATION olake_publication FOR ALL TABLES
WITH (publish = 'insert,update');
-- For partitioned tables (PostgreSQL 13+): publish changes via the partition root
CREATE PUBLICATION olake_publication FOR ALL TABLES
WITH (publish = 'insert,update,delete,truncate', publish_via_partition_root = true);
Verify Publication:
-- List all publications
SELECT * FROM pg_publication;
-- View tables in a publication
SELECT * FROM pg_publication_tables WHERE pubname = 'olake_publication';
Add Tables to Existing Publication:
-- Add a single table
ALTER PUBLICATION olake_publication ADD TABLE public.new_table;
-- Add multiple tables
ALTER PUBLICATION olake_publication ADD TABLE public.table4, public.table5;
-- Remove a table
ALTER PUBLICATION olake_publication DROP TABLE public.old_table;
5. Create Replication Slot
Set up the replication slot for OLake to consume changes:
Use the exact replication slot name that you configure in OLake UI or CLI. The slot name must match the replication_slot parameter in your OLake source configuration.
Best practices:
- Create the publication before creating the replication slot
- Create the slot in the same database that OLake will connect to (slots are per-database)
- Use a unique slot name per OLake connection; slot names cannot start with a number
-- Replace 'your_olake_slot_name' with the slot name from your OLake configuration
SELECT pg_create_logical_replication_slot('your_olake_slot_name', 'pgoutput');
Example with common slot names:
-- If your OLake source config uses "olake_slot"
SELECT pg_create_logical_replication_slot('olake_slot', 'pgoutput');
-- If your OLake source config uses "postgres_slot"
SELECT pg_create_logical_replication_slot('postgres_slot', 'pgoutput');
Verify Slot Creation:
SELECT * FROM pg_replication_slots WHERE slot_name = 'your_olake_slot_name';
6. Test Logical Decoding
Verify that CDC is working correctly with pgoutput:
-- Create test data
CREATE TABLE IF NOT EXISTS test_table (
id SERIAL PRIMARY KEY,
data TEXT,
created_at TIMESTAMP DEFAULT CURRENT_TIMESTAMP
);
-- Set replica identity
ALTER TABLE test_table REPLICA IDENTITY DEFAULT;
-- Add table to publication
ALTER PUBLICATION olake_publication ADD TABLE test_table;
-- Insert test data
INSERT INTO test_table (data) VALUES ('PostgreSQL pgoutput CDC test - INSERT');
Check for changes using pg_logical_slot_peek_binary_changes:
SELECT *
FROM pg_logical_slot_peek_binary_changes(
'olake_slot', NULL, NULL,
'proto_version','1',
'publication_names','olake_publication'
);
Expected Results:
✅ Success: Returns binary change records from pgoutput
❌ Permission denied: Check user has REPLICATION role
❌ Publication not found: Verify publication exists and contains tables
❌ Slot not found: Check replication slot was created with pgoutput plugin
❌ No changes returned: Verify table has replica identity configured and is in the publication
Drop test slots after verification: SELECT pg_drop_replication_slot('test_slot');
Now that you have configured the database and created the CDC user, you can add the PostgreSQL source in OLake to build an ELT pipeline to Iceberg or Parquet. See the PostgreSQL connector overview for a high-level walkthrough.
Troubleshooting
-
Permission Errors: If you see
ERROR: permission denied to create replication slotor the connector logs an authentication error for replication, it means the user lacks replication privileges. Use the master user or ensure your user has therds_superuser(or at leastrds_replication) role. On Aurora, using the defaultpostgresuser is simplest, since it already has the needed privileges. -
Plugin Not Found: If you get an error like
ERROR: could not access file "pgoutput": No such file or directorywhen creating a slot, ensure PostgreSQL version is 10+ andrds.logical_replicationis applied. Re-check the parameter group (step 1) and reboot. -
No Changes Captured: If the slot exists and the connector is running but no changes are coming through:
-
Verify that
wal_levelis indeed logical on the Aurora instance (you canSHOW wal_level;to confirm). -
Ensure the table you are changing is in a replication-enabled database (in Postgres, logical decoding is per database). The connector should connect to the specific database where the changes occur.
-
Check the replication slot statistics: run
SELECT * FROM pg_replication_slots;. If the slot’sactivecolumn isf(false), the connector isn’t actually connected. Ifactiveistbutrestart_lsnisn’t advancing, the connector might be stuck or not processing events. -
OLake uses the native pgoutput plugin and requires a configured publication.
-
-
WAL Retention and Bloat: Logical replication slots will retain WAL segments on the cluster until they are consumed. If a slot is not actively read, the WAL will pile up and can eventually fill storage. On Aurora, monitor the “Write-Ahead Log consumption” or free storage. If your connector falls far behind or is stopped, you may see disk usage grow or even an error in the database logs about WAL retention. To resolve, resume consumption or if the slot is no longer needed, drop it. Always drop unused logical slots
-
Replica Identity Issues: PostgreSQL requires a primary key (or other replica identity) on tables to capture UPDATE/DELETE properly. If a table has no primary key and you have not set
REPLICA IDENTITY FULL, deleting or updating rows can cause logical decoding to skip or not output those changes. If you see messages about “no replica identity” or missing information for some changes, consider settingREPLICA IDENTITY FULLon those tables:ALTER TABLE my_table REPLICA IDENTITY FULL;This will include the full before-image of rows in WAL for tables without a primary key, ensuring pgoutput can output the changes.
-
Network Connectivity: If the CDC application cannot connect at all, check the Aurora cluster’s security group. The Aurora instance must allow inbound connections from your CDC client (open the PostgreSQL port, default 5432).
With Aurora PostgreSQL configured as above, you have logical decoding streaming changes via pgoutput. This sets the stage for feeding those changes into downstream systems (data warehouses, Kafka, etc.) in a reliable, low-latency manner.